Glycomic Elucidation and Annotation Tool, GELATO, is a semi-automated MS/MS annotation tool that rapidly matches hundreds of experimental MS/MS spectra with theoretical glycan fragments from highly curated default glycan databases known as SweetyN and SweetyO. These databases are derived from the human curated Glycan Ontology (GlycO), developed at the CCRC and curated by Michael Tiemeyer and Kazuhiro Aoki. An overview of the currently curated structures is available on the status page of the curation tool Qrator. The structures present in the databases are fragmented in silico using Glycoworkbench program libraries and matched against experimental spectra to identify candidate structures. GELATO reports matches as the number of peaks matched and % of the total intensity of the peaks in the experimental spectrum. GELATO is capable of considering multiple charge states for each m/z reported and neutral losses including under-permethylation and loss of water.
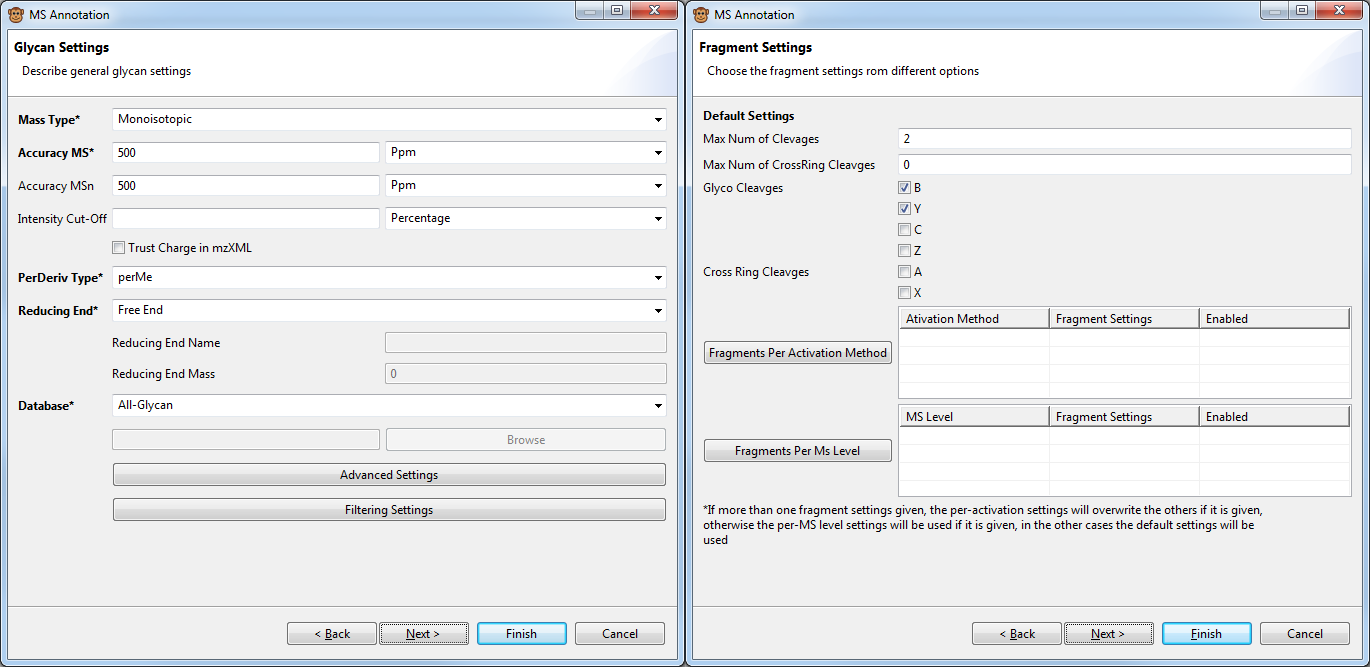
Figure 1: Screenshot of GELATO fragmentation settings dialogs. Left dialog: General annotation settings including accuracy, glycan preparation (derivatisation, reducing end type) and database to be used for annotation. Right dialog: Fragmentation settings. IN the upper half the default fragmentation settings (number of fragments, allowed fragment type) can be defined. In the lower section special fragmentation settings per MS level or activation method can be defined an will be used instead of the default settings for corresponding spectra.
In addition to the annotation of experimental GRITS extracts key information from the annotation results of either GELATO along with information contained in the RAW data file produced by the mass spectrometer and combines this information into a sortable table that reports the following information:
- m/z, z, signal intensity, and scan number for each spectra
- GELATO match statistics peak count and % intensity coverage
- A cartoon representation, generated by GlycoWorkbench, along with the corresponding exact mass and the associated sequence string
- A set of descriptors for the assigned structure including: # Charges, # Hex, # HexNAc, # NeuAc, # NeuGc, # Fuc, # Sulfate, # Phosphate, # HexA, # Other, # Branches, # Gal-Gal disaccharide groups, # Non core Fuc, # LacNAc groups, # LacDiNAc groups, Bisecting GlcNAc present?, Core Fuc present?, Polysialic acid present?, and Glycan Type (High-Man, Bisecting, Hybrid, Complex)
- Signal intensities detected for user defined specific diagnostic MS/MS fragment ions (e.g., m/z=660 reports a monofucosylated LacNAc terminal), providing a diagnostic for structural validation and relative comparisons between samples
After the calculation the results are stored as part of the project and can be displayed and manually post processed. The tool presents the annotation results in a table format and users can remove automatic generated annotations based on there expertise and refine the automatic generated results.
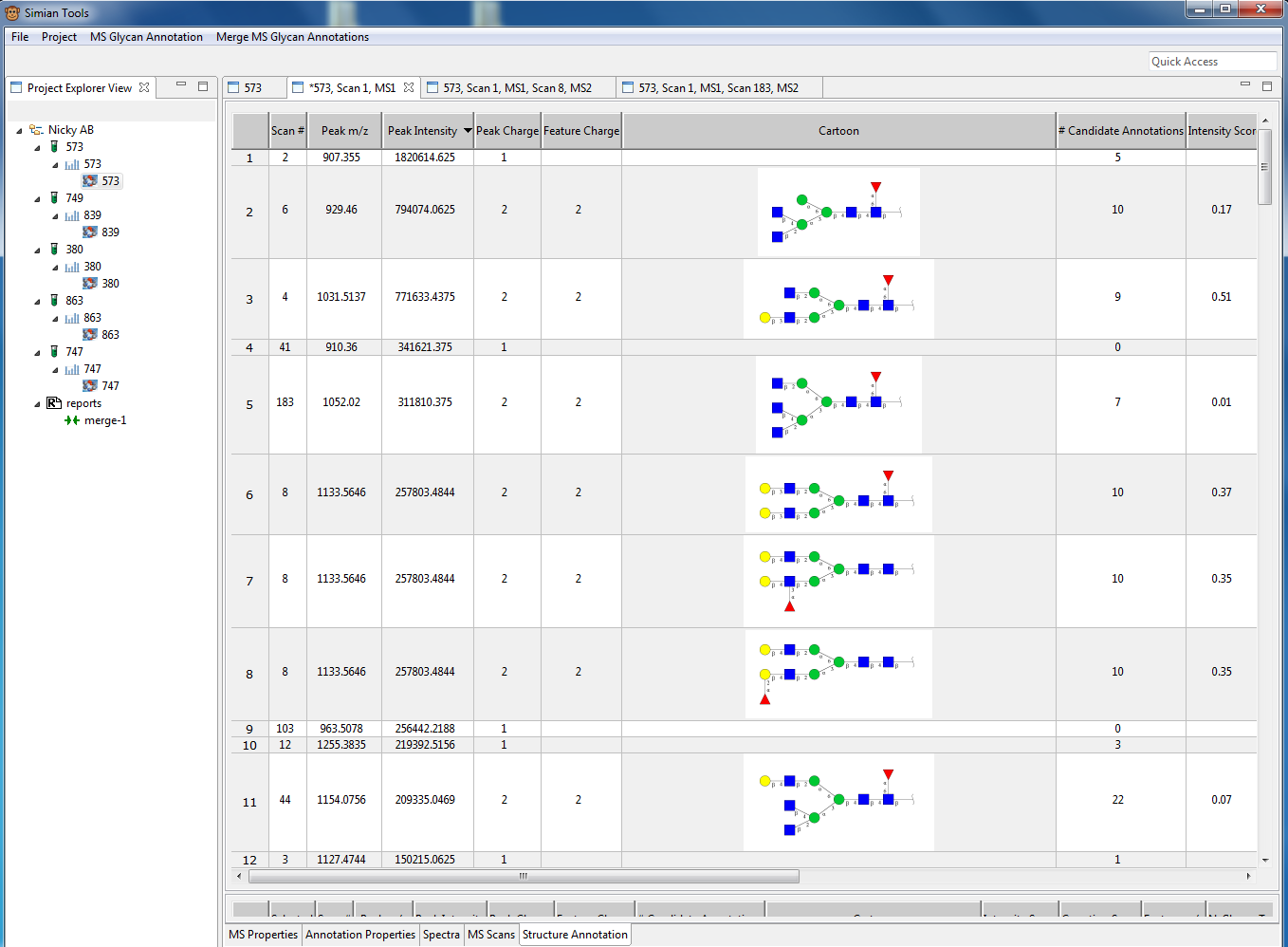
Figure 2: Screenshot of the MS1 annotation of an experiment. The table shows the glycan in cartoon representation together with peak information (m/z, intensity) and additional calculated meta data. By single clicking on peak/structures in the table a list of alternative annotation is shown and users can add or remove annotations. If a MSn scan is available for the peak user can open the scan by double clicking on the row.
In the MS profile perspective (see Figure 2) each peak is annotated with theoretical candidate structures found in the database utilized by GELATO. By selecting a row all candidates for a peak are shown and a user can manually deselect structures. Alternative the user can double click a row and see the MS2 annotations if the MS1 peak was select as a precursor for a MS2 scan by the instrument.